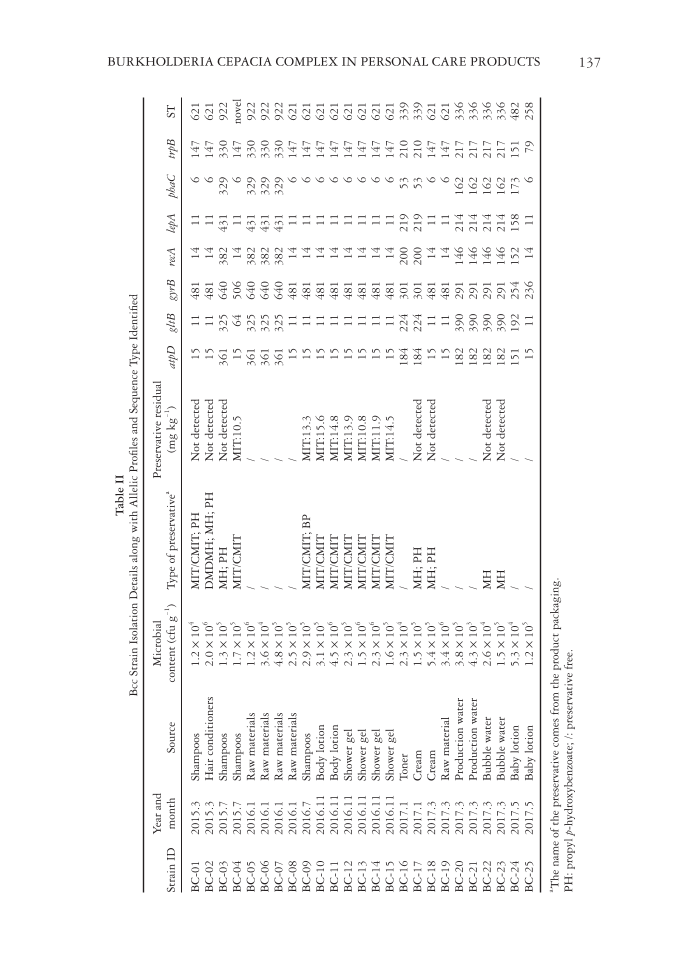
Table II Bcc Strain Isolation Details along with Allelic Profi les and Sequence Type Identifi ed Strain ID Year and month Source Microbial content (cfu g)-1 Type of preservative a Preservative residual (mg kg-1) atpD gltB gyrB recA lepA phaC trpB ST BC-01 2015.3 Shampoos 1.2 × 10 4 MIT/CMIT PH Not detected 15 11 481 14 11 6 147 621 BC-02 2015.3 Hair conditioners 2.0 × 10 6 DMDMH MH PH Not detected 15 11 481 14 11 6 147 621 BC-03 2015.7 Shampoos 1.3 × 10 5 MH PH Not detected 361 325 640 382 431 329 330 922 BC-04 2015.7 Shampoos 1.7 × 10 5 MIT/CMIT MIT:10.5 15 64 506 14 11 6 147 novel BC-05 2016.1 Raw materials 1.2 × 10 6 / / 361 325 640 382 431 329 330 922 BC-06 2016.1 Raw materials 3.6 × 10 4 / / 361 325 640 382 431 329 330 922 BC-07 2016.1 Raw materials 4.8 × 10 5 / / 361 325 640 382 431 329 330 922 BC-08 2016.1 Raw materials 2.5 × 10 5 / / 15 11 481 14 11 6 147 621 BC-09 2016.7 Shampoos 2.9 × 10 5 MIT/CMIT BP MIT:13.3 15 11 481 14 11 6 147 621 BC-10 2016.11 Body lotion 3.1 × 10 5 MIT/CMIT MIT:15.6 15 11 481 14 11 6 147 621 BC-11 2016.11 Body lotion 4.5 × 10 6 MIT/CMIT MIT:14.8 15 11 481 14 11 6 147 621 BC-12 2016.11 Shower gel 2.3 × 10 5 MIT/CMIT MIT:13.9 15 11 481 14 11 6 147 621 BC-13 2016.11 Shower gel 1.5 × 10 6 MIT/CMIT MIT:10.8 15 11 481 14 11 6 147 621 BC-14 2016.11 Shower gel 2.3 × 10 6 MIT/CMIT MIT:11.9 15 11 481 14 11 6 147 621 BC-15 2016.11 Shower gel 1.6 × 10 5 MIT/CMIT MIT:14.5 15 11 481 14 11 6 147 621 BC-16 2017.1 Toner 2.3 × 10 4 / / 184 224 301 200 219 53 210 339 BC-17 2017.1 Cream 1.5 × 10 5 MH PH Not detected 184 224 301 200 219 53 210 339 BC-18 2017.3 Cream 5.4 × 10 5 MH PH Not detected 15 11 481 14 11 6 147 621 BC-19 2017.3 Raw material 3.4 × 10 6 / / 15 11 481 14 11 6 147 621 BC-20 2017.3 Production water 3.8 × 10 5 / / 182 390 291 146 214 162 217 336 BC-21 2017.3 Production water 4.3 × 10 3 / / 182 390 291 146 214 162 217 336 BC-22 2017.3 Bubble water 2.6 × 10 4 MH Not detected 182 390 291 146 214 162 217 336 BC-23 2017.3 Bubble water 1.5 × 10 5 MH Not detected 182 390 291 146 214 162 217 336 BC-24 2017.5 Baby lotion 5.3 × 10 4 / / 151 192 254 152 158 173 151 482 BC-25 2017.5 Baby lotion 1.2 × 10 5 / / 15 11 236 14 11 6 79 258 a The name of the preservative comes from the product packaging. PH: propyl p /: preservative free. BURKHOLDERIA CEPACIA COMPLEX IN PERSONAL CARE PRODUCTS 137
JOURNAL OF COSMETIC SCIENCE 138 Table III List of Primers Used in the MLST Locus Amplifi cation Gene name Size (bp) of fragment analyzed Locus primer (5′→3′) Locus primer (5′→3′) atpD 443 GTTCATCTGGCCGTACAC AACTGACGCTCGAAGTCC gltB 400 CTTCTTCTTCGTCGCCGA TTGCCGACGTAGTCGTTG gyrB 454 ATCGTGATGACCGAGCTG CGTTGTAGCTGTCGTTCC recA 393 TGACCGCCGAGAAGAGCAA GACCGAGTCGATGACGAT lepA 397 GGCATCAAGGAACTGACG CTGCGGCATGTACAGGTT phaC 385 AGACGGCTTCAAGGTGGT ACACGGTGTTGACCGTCA trpB 301 CTGGGTCACGAACATGGA CCGAATGCGTCTCGATGA analyses were conducted using genetic-distance–based neighbor-joining algorithms within MEGA version 4.0 software package (Tokyo, Japan). MLST LOCI AMPLIFICAT ION AND SEQUENCING MLST was performed b y sequencing the following housekeeping genes: ATP synthase beta chain (atpD), glutamate synthase large subunit (gltB), DNA gyrase subunit B (gyrB), recombinase A (recA), GTP-binding protein (lepA), acetoacetyl-CoA reductase (phaC), and tryptophan synthase subunit B (trpB) according to the previously published method available at www.pubmlst.org/Bcc. Primer sequences used for the MLST locus amplifi ca- tion are listed in Table III. Genomic DNA was isol ated using the genomic DNA extraction kit (TaKaRa) mentioned previously, according to the manufacturer’s instructions for bacterial cells. Amplifi cation of MLST loci was performed in 50 μl PCR volumes containing 2× EasyTaq® PCR Super- Mix (TransGen Biotech, Beijing, China), 0.4 μM primers, DNA templates, and double distilled water. Amplifi cation was performed with the Eppendorf Mastercycler® 5,330 using the following cycling conditions: initial denaturation for 2 min at 94°C followed by 30 cycles of 1 min at 94°C, 30 s at an annealing temperature of 58°C, and 2 min at 72°C, followed by a fi nal extension step of 5 min at 72°C. The reaction products were separated and detected on an ABI PRISM Genetic Analyzer 3100 (Applied Biosystems). The sequences from both strands of a given locus of the same isolate were aligned, trimmed to the desired length, and edited using the SeqMan II program from the Laser- gene software package. The specifi c ST of the analyzed strains was determined using the B. cepacia complex multilocus sequence typing website (http://pubmlst.org/Bcc/) devel- oped by Keith Jolley at the University of Oxford (England, United Kingdom) (39). ANTIMICROBIAL SUSCEPTIBILITY T ESTING Determination of the MIC. Mini mum inhibitory concentrati ons (MICs) were determined on a panel of 25 strains illustrating the diverse origins of the collection, broth microdilution using 96-well microplate. First, preservative mother solution was prepared according to the concentrations from Table IV and diluted 10 times, and then a double dilution up to six different dilutions (Table IV) was made. For each bacterial isolate to be tested, 100 μl
Purchased for the exclusive use of nofirst nolast (unknown) From: SCC Media Library & Resource Center (library.scconline.org)