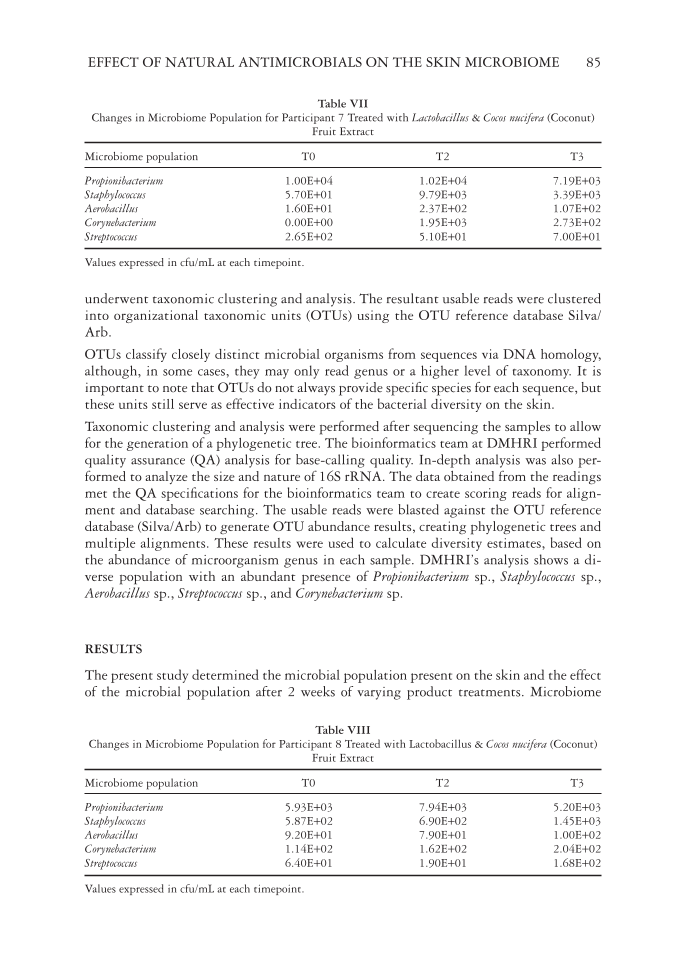
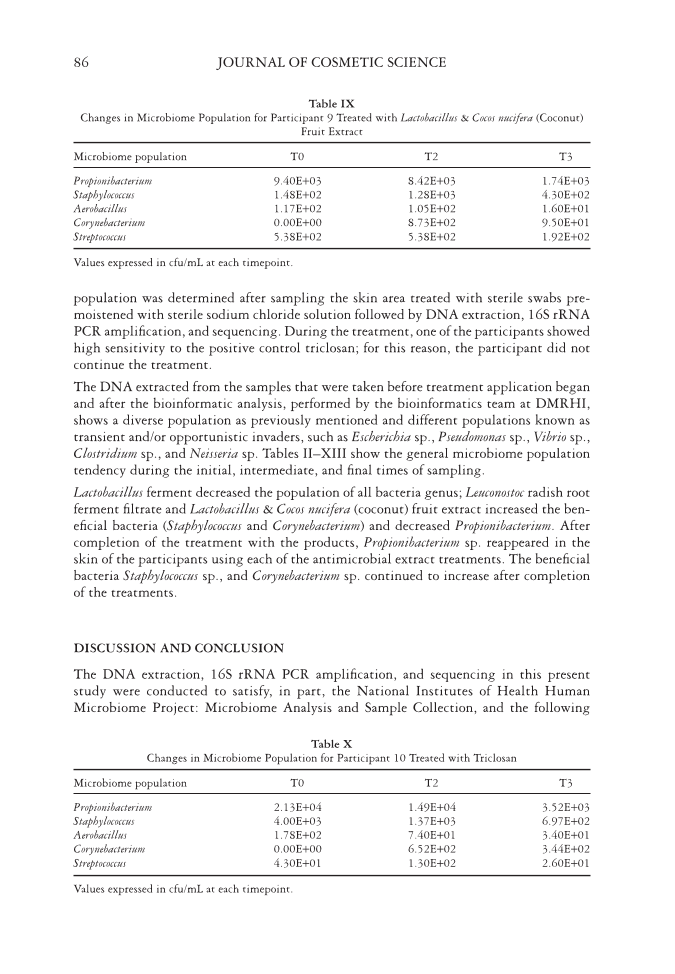
EFFECT OF NATURAL ANTIMICROBIALS ON THE SKIN MICROBIOME 85 underwent taxonomic clustering and analysis. The resultant usable reads were clustered into organizational taxonomic units (OTUs) using the OTU reference database Silva/ Arb. OTUs classify closely distinct microbial organisms from sequences via DNA homology, although, in some cases, they may only read genus or a higher level of taxonomy. It is important to note that OTUs do not always provide specifi c species for each sequence, but these units still serve as effective indicators of the bacterial diversity on the skin. Taxonomic clustering and analysis were performed after sequencing the samples to allow for the generation of a phylogenetic tree. The bioinformatics team at DMHRI performed quality assurance (QA) analysis for base-calling quality. In-depth analysis was also per- formed to analyze the size and nature of 16S rRNA. The data obtained from the readings met the QA specifi cations for the bioinformatics team to create scoring reads for align- ment and database searching. The usable reads were blasted against the OTU reference database (Silva/Arb) to generate OTU abundance results, creating phylogenetic trees and multiple alignments. These results were used to calculate diversity estimates, based on the abundance of microorganism genus in each sample. DMHRI’s analysis shows a di- verse population with an abundant presence of Propionibacterium sp., Staphylococcus sp., Aerobacillus sp., Streptococcus sp., and Corynebacterium sp. RESULTS The present study determined the microbial population present on the skin and the effect of the microbial population after 2 weeks of varying product treatments. Microbiome Table VII Changes in Microbiome Population for Participant 7 Treated with Lactobacillus & Cocos nucifera (Coconut) Fruit Extract Microbiome population T0 T2 T3 Propionibacterium 1.00E+04 1.02E+04 7.19E+03 Staphylococcus 5.70E+01 9.79E+03 3.39E+03 Aerobacillus 1.60E+01 2.37E+02 1.07E+02 Corynebacterium 0.00E+00 1.95E+03 2.73E+02 Streptococcus 2.65E+02 5.10E+01 7.00E+01 Values expressed in cfu/mL at each timepoint. Table VIII Changes in Microbiome Population for Participant 8 Treated with Lactobacillus & Cocos nucifera (Coconut) Fruit Extract Microbiome population T0 T2 T3 Propionibacterium 5.93E+03 7.94E+03 5.20E+03 Staphylococcus 5.87E+02 6.90E+02 1.45E+03 Aerobacillus 9.20E+01 7.90E+01 1.00E+02 Corynebacterium 1.14E+02 1.62E+02 2.04E+02 Streptococcus 6.40E+01 1.90E+01 1.68E+02 Values expressed in cfu/mL at each timepoint.
JOURNAL OF COSMETIC SCIENCE 86 population was determined after sampling the skin area treated with sterile swabs pre- moistened with sterile sodium chloride solution followed by DNA extraction, 16S rRNA PCR amplifi cation, and sequencing. During the treatment, one of the participants showed high sensitivity to the positive control triclosan for this reason, the participant did not continue the treatment. The DNA extracted from the samples that were taken before treatment application began and after the bioinformatic analysis, performed by the bioinformatics team at DMRHI, shows a diverse population as previously mentioned and different populations known as transient and/or opportunistic invaders, such as Escherichia sp., Pseudomonas sp., Vibrio sp., Clostridium sp., and Neisseria sp. Tables II–XIII show the general microbiome population tendency during the initial, intermediate, and fi nal times of sampling. Lactobacillus ferment decreased the population of all bacteria genus Leuconostoc radish root ferment fi ltrate and Lactobacillus & Cocos nucifera (coconut) fruit extract increased the ben- efi cial bacteria (Staphylococcus and Corynebacterium) and decreased Propionibacterium. After completion of the treatment with the products, Propionibacterium sp. reappeared in the skin of the participants using each of the antimicrobial extract treatments. The benefi cial bacteria Staphylococcus sp., and Corynebacterium sp. continued to increase after completion of the treatments. DISCUSSION AND CONCLUSION The DNA extraction, 16S rRNA PCR amplifi cation, and sequencing in this present study were conducted to satisfy, in part, the National Institutes of Health Human Microbiome Project: Microbiome Analysis and Sample Collection, and the following Table IX Changes in Microbiome Population for Participant 9 Treated with Lactobacillus & Cocos nucifera (Coconut) Fruit Extract Microbiome population T0 T2 T3 Propionibacterium 9.40E+03 8.42E+03 1.74E+03 Staphylococcus 1.48E+02 1.28E+03 4.30E+02 Aerobacillus 1.17E+02 1.05E+02 1.60E+01 Corynebacterium 0.00E+00 8.73E+02 9.50E+01 Streptococcus 5.38E+02 5.38E+02 1.92E+02 Values expressed in cfu/mL at each timepoint. Table X Changes in Microbiome Population for Participant 10 Treated with Triclosan Microbiome population T0 T2 T3 Propionibacterium 2.13E+04 1.49E+04 3.52E+03 Staphylococcus 4.00E+03 1.37E+03 6.97E+02 Aerobacillus 1.78E+02 7.40E+01 3.40E+01 Corynebacterium 0.00E+00 6.52E+02 3.44E+02 Streptococcus 4.30E+01 1.30E+02 2.60E+01 Values expressed in cfu/mL at each timepoint.
Purchased for the exclusive use of nofirst nolast (unknown) From: SCC Media Library & Resource Center (library.scconline.org)