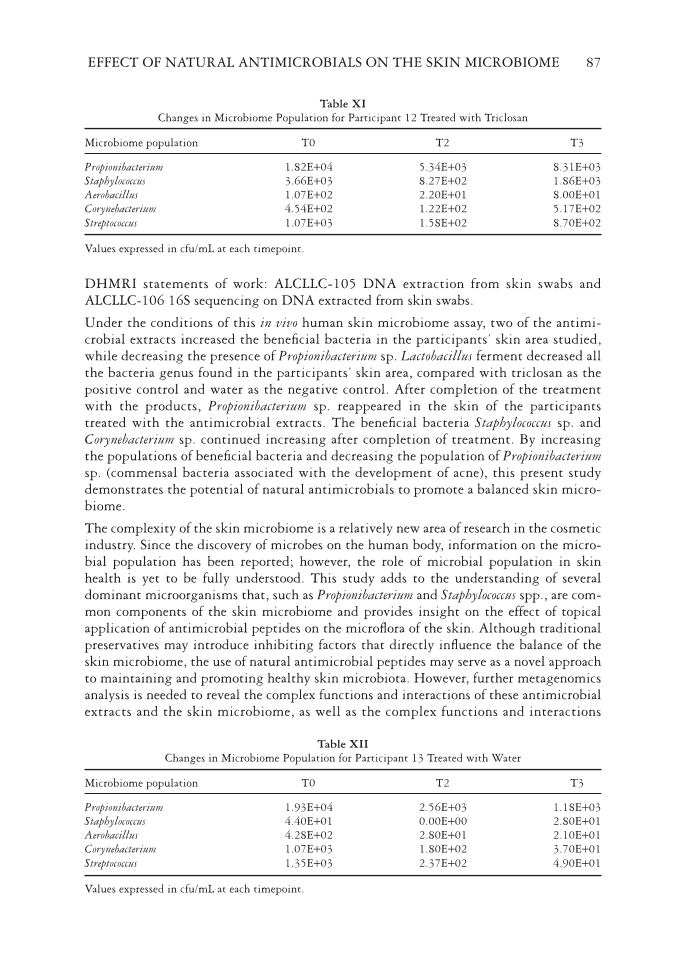
EFFECT OF NATURAL ANTIMICROBIALS ON THE SKIN MICROBIOME 87 Table XI Changes in Microbiome Population for Participant 12 Treated with Triclosan Microbiome population T0 T2 T3 Propionibacterium 1.82E+04 5.34E+03 8.31E+03 Staphylococcus 3.66E+03 8.27E+02 1.86E+03 Aerobacillus 1.07E+02 2.20E+01 8.00E+01 Corynebacterium 4.54E+02 1.22E+02 5.17E+02 Streptococcus 1.07E+03 1.58E+02 8.70E+02 Values expressed in cfu/mL at each timepoint. Table XII Changes in Microbiome Population for Participant 13 Treated with Water Microbiome population T0 T2 T3 Propionibacterium 1.93E+04 2.56E+03 1.18E+03 Staphylococcus 4.40E+01 0.00E+00 2.80E+01 Aerobacillus 4.28E+02 2.80E+01 2.10E+01 Corynebacterium 1.07E+03 1.80E+02 3.70E+01 Streptococcus 1.35E+03 2.37E+02 4.90E+01 Values expressed in cfu/mL at each timepoint. DHMRI statements of work: ALCLLC-105 DNA extraction from skin swabs and ALCLLC-106 16S sequencing on DNA extracted from skin swabs. Under the conditions of this in vivo human skin microbiome assay, two of the antimi- crobial extracts increased the benefi cial bacteria in the participants’ skin area studied, while decreasing the presence of Propionibacterium sp. Lactobacillus ferment decreased all the bacteria genus found in the participants’ skin area, compared with triclosan as the positive control and water as the negative control. After completion of the treatment with the products, Propionibacterium sp. reappeared in the skin of the participants treated with the antimicrobial extracts. The benefi cial bacteria Staphylococcus sp. and Corynebacterium sp. continued increasing after completion of treatment. By increasing the populations of benefi cial bacteria and decreasing the population of Propionibacterium sp. (commensal bacteria associated with the development of acne), this present study demonstrates the potential of natural antimicrobials to promote a balanced skin micro- biome. The complexity of the skin microbiome is a relatively new area of research in the cosmetic industry. Since the discovery of microbes on the human body, information on the micro- bial population has been reported however, the role of microbial population in skin health is yet to be fully understood. This study adds to the understanding of several dominant microorganisms that, such as Propionibacterium and Staphylococcus spp., are com- mon components of the skin microbiome and provides insight on the effect of topical application of antimicrobial peptides on the microfl ora of the skin. Although traditional preservatives may introduce inhibiting factors that directly infl uence the balance of the skin microbiome, the use of natural antimicrobial peptides may serve as a novel approach to maintaining and promoting healthy skin microbiota. However, further metagenomics analysis is needed to reveal the complex functions and interactions of these antimicrobial extracts and the skin microbiome, as well as the complex functions and interactions
JOURNAL OF COSMETIC SCIENCE 88 Table XIII Changes in Microbiome Population for Participant 15 Treated with Water Microbiome population T0 T2 T3 Propionibacterium 2.07E+03 5.09E+03 4.12E+02 Staphylococcus 2.00E+00 7.00E+00 0.00E+00 Aerobacillus 6.10E+01 1.28E+02 1.20E+01 Corynebacterium 1.38E+02 3.51E+02 1.80E+01 Streptococcus 1.30E+02 1.50E+01 0.00E+00 Values expressed in cfu/mL at each timepoint. between the microorganisms of the skin microbiome. Further research in this area could provide more comprehensive approaches to the development of topical products that consider the integral contributions of skin microbiome. REFERENCES (1) D. Erturk-Hasdemir and D. Kasper, Resident commensals shaping immunity, Curr. Opin. Immunol., 25, 450–455 (2013). (2) M. Danaher, D. Scholz, E. Segura, and M. Darley, Natural vs. synthetic antimicrobials and HDAC as an indicator of microfl ora health, Cosmet. Toiletries, 130, 22–34 (2015). (3) A. Cogen, V. Nizet, and R. Gallo, Skin microbiota: a source of disease or defence? Br. J. Dermatol., 158, 442–455 (2008). (4) M. R. Yeaman and N. Y. Yount, Mechanisms of antimicrobial peptide action and resistance, Pharmacol. Rev., 55, 27–55 (2003). (5) T. Prince, A. J. McBain, C. A. O’Neill, Lactobacillus reuteri protects epidermal keratinocytes from Staph- ylococcus aureus-induced cell death by competitive exclusion, Appl. Environ. Microbiol., 78, 5119–5126 (2012). (6) N. Izawa and T. Sone, “Cosmetic ingredients fermented by lactic acid bacteria,” in Microbial Production: From Genome Design to Cell Engineering, H. Anazawa and S. Shimizu. Eds. (Springer, Tokyo, Japan, 2014), pp. 233–242. (7) J. Ouyang, Z. Pei, L. Lutwick, S. Dalal, L. Yang, N. Cassai, K. Sandhu, B. Hanna, R. Wieczorek, M. Bluth, and M. Pincus, Case report: Paenibacillus thiaminolyticus: a new cause of human infection, induc- ing bacteremia in a patient on hemodialysis, Ann. Clin. Lab. Sci., 38, 393–400 (2008). (8) J. M. Janda and L. Abott, 16S rRNA gene sequencing for bacterial identifi cation in the diagnostic laboratory: pluses, perils, and pitfalls, J. Clin. Microbiol., 45, 2761–2764 (2007). (9) K. H. Wilson, R. B. Blitchinton, and R.C. Greene, Amplifi cation of bacterial 16S ribosomal DNA with polymerase chain reaction, J. Clin. Microbiol., 28, 1942–1946 (1990).
Purchased for the exclusive use of nofirst nolast (unknown) From: SCC Media Library & Resource Center (library.scconline.org)