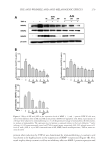
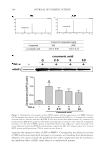
JOURNAL OF COSMETIC SCIENCE 250 Phylum Class Genus Number Gammaproteobacteria Cardiobacterium 10 Enterobacter 7 Escherichia 13 Salmonella 2 Trabulsiella 1 Halomonas 5 Oceanospirillales_uc_g 1 Haemophilus 712 Pasteurellaceae_uc 16 Acinetobacter 5 Moraxella 27 Moraxellaceae_uc 2 Pseudomonadaceae_uc 5 Pseudomonas 190 Pseudomonadales_uc_g 1 Lysobacter 4 Xanthomonadaceae_uc 1 Xanthomonadales_uc_g 1 Saccharibacteria_TM7 Saccharimonas_c Saccharimonas 9 Verrucomicrobia Verrucomicrobiae Pedosphaera_f_uc 2 Unkown 102 Total 19,863 Table I Continued Repeated use of cosmetics can cause microbial contamination and infection (14). In general, cosmetics should be free from the following bacterial species: S. aureus, E. coli, Salmonella spp., C. albicans, Clostridium spp., and P. aeruginosa (3). Staphylococcus and Pseudomonas were identifi ed in this experiment, whereas pathogenic bacteria such as Escherichia and Salmonella were found to spread through the fecal-oral route. In addition, oral bacteria associated with disease, such as Actinomyces, Porphyromonas, and Fusobacterium were found. Pathogenic bacteria such as Corynebacterium and Neisseria were also found in this study. In particular, Corynebacterium diphtheriae of the Corynebacterium genus is the cause of diphtheria (15). Neisseria meningitidis and Neisseria gonorrhoeae of the Neisseria genus cluster on the mucosal surfaces of humans. The former is known to cause sepsis and meningitis, whereas the latter causes gonorrhea (16). As our sequencing was not deep enough to identify the particular species present, additional studies will be needed to determine whether these pathogenic bacteria are present in lipstick. In addition, a genus was considered pathogenic if any one of its members were known pathogens. Therefore, the actual proportion of pathogenic bacteria might be lower than reported here. Previous studies examining bacterial contamination of lipstick using classical bacterial cultures have reported contamination by various species. Onondaga et al. conducted a Gram stain and biochemical test on 20 lipstick samples (3) and observed contamination by S. aureus and C. albicans. Sawant et al. examined 12 lipstick samples for contamination before and
BACTERIAL CONTAMINATION OF LIPSTICK 251 after use (4). Proteus, Providencia, Morganella, Staphylococcus, and Pseudomonas were detected using 16S rDNA sequencing, Gram stain, and biochemical characterization. Although these studies are limited to culturable bacteria, they have nonetheless detected the pres- ence of various genera of bacteria on lipstick. Preservatives are commonly used to ensure the stability and safety of cosmetic products (14,17). Various preservatives have been used to maintain low levels of microorganism contamination and to increase the shelf life of lipstick (4,18). Air contact following open- ing of the cap has been reported to increase microbial contamination, although preserva- tives possess suffi cient antimicrobial activity to maintain product safety (4). In our study, 20 lipstick samples were plated on a blood agar plate and cultured in a 5% CO2 incubator for 48 h. Live bacteria were found in seven of 20 samples (data not shown). These results suggest that the antiseptics contained in lipstick may be present in insuffi cient concen- trations for antimicrobial activity resulting in contamination. In this study, we investigated the diversity of contaminating bacteria in lipstick using pyrosequencing. We detected a wider diversity of contaminating bacteria compared with previous studies. Lipstick comes into direct contact with the mouth, and because it is reused, microorganisms can infect the skin as well. We suggest that consumers should use products that inhibit the growth of contaminating bacteria on their cosmetics, and that the types of preservatives as well as their concentrations should be optimized. REFERENCES (1) J. Behra van, F. Bazzaz, and P. Malaekeh, Survey of bacteriological contamination of cosmetic creams in Iran (2000). Int. J. Dermatol., 44, 482–485 (2005). (2) R. Campa na, C. Scesa, V. Patrone, E. Vittoria, and W. Baffone, Microbiological study of cosmetic products during their use by consumers: Health risk and effi cacy of preservative systems. Lett. Appl. Microbiol., 43, 301–306 (2006). Figure 1. Dive rsity of the bacterial community present in lipstick samples.
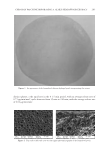
Purchased for the exclusive use of nofirst nolast (unknown) From: SCC Media Library & Resource Center (library.scconline.org)