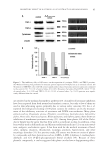
INHIBITORY MECHANISM OF RED GLOBE AMARANTH ON TYROSINASE 107 assumed to be band K (210–250 nm) and band B (260–300 nm) of a benzene ring. The maximum absorptions of VA remained the same after the addition of excess Cu2+, whereas the absorption maxima shifted to 249 and 285 nm after the addition of tyrosinase. These results illustrate that the VA can interact with tyrosinase however, chelation interaction between VA and the dicopper copper ion of the enzyme is not included (30). STEREOSCOPIC STRUCTURAL HOMOLOGY ANALYSIS The conformations of VA docked onto tyrosinase were investigated to determine the mechanism of inhibition of mushroom tyrosinase by VA. Because a three-dimensional structure of mushroom tyrosinase has not yet been reported, we used crystal structure of tyrosinase from B. megaterium as a model (Figure 4, dark) (23). Before the molecular dock- ing was performed, the structure of mushroom tyrosinase (TyrMu) (Figure 4, light) was predicted according to its primary sequence (GenBank ID: CAC82195.1) from the Phyre Server (28) for comparison. We superimposed the predicted structure of TyrMu with the crystal structure of TyrBm using the secondary structure matching superimposition pro- gram Coot (29). The superimposition results of the overall structures and the active sites are shown in Figure 4. The predictions indicated that the catalytic core domains of the two three-dimensional structures are highly conserved. Therefore, it is reasonable to choose the active center of TyrBm to serve as our structural model. MOLECULAR DOCKING ANALYSIS The molecular simulations of the interaction between VA and tyrosinase are shown in Figure 5. VA was bound by interactions with residues Glu192 and Gly213 (Glu195 and Gly216 in TyrBm) at the entrance to the active center and His57 (His60 in TyrBm) in the active center (Figure 5A). Thus, the conformation of the active pocket may be changed by VA as a result of the interaction with adjacent residues (23). Furthermore, we Figure 4. Superimposition of TyrBm (dark) and the prediction of the mushroom tyrosinase structure (light). The two balls were dicopper center of tyrosinase.
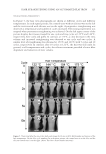
JOURNAL OF COSMETIC SCIENCE 108 superimposed VA with substrates in PyMOL (Figure 5B). The overlap prediction indi- cates that the three-dimensional position occupied by VA is almost the same as that of the substrates at the tyrosinase active core domain. Thus, VA obstructs the correct orientation of the substrates at the catalytic center, which suggests one possible mechanism of inhibi- tion (23). Sendovski et al. suggested that His57 (His60 in TyrBm) is responsible for the deprotonation of the monophenol substrate (23). The interaction of VA with His57 im- plies that VA may have a greater effect on monophenolase activity than on diphenolase activity, which is consistent with the results described above. For the modeling of molecular docking using Rosetta, the value of the total score is re- lated to the free energy of the receptor–ligand complex. The total scores were −936.57, −934.99, and −936.46 for the modeling of docking with VA, L -tyrosine, and L -dopa, re- spectively. Thus, the tyrosinase–VA complex exhibited greater stability compared with that of the substrate complexes. Thus, VA may bind to tyrosinase more easily and more strongly than monophenol and diphenol substrates. As a result, VA could inhibit tyrosi- nase activity in the presence of substrates, as shown in Table IV. In addition, because the tyrosinase–L-tyrosine complex is the least stable, the competition by the inhibitor may be stronger for monophenol substrate, which may be another reason that the monophenolase activity is lower than the diphenolase activity in the presence of VA. Figure 5. Suggested docking model for VA and tyrosinase. (A) Interaction of VA and tyrosinase in the dock- ing model. (B) Superimposition of VA with L -tyrosine and L -dopa.
Purchased for the exclusive use of nofirst nolast (unknown) From: SCC Media Library & Resource Center (library.scconline.org)