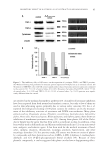
MODULATION OF CELLULAR SENESCENCE 81 (14,17,19), improved collagen synthesis (15,18,19), and increased blood vessel devel- opment (17), appear to be principally related to unique proteins and low molecular weight peptides that are enzymatically manufactured in the growing yeast (17,18). Undoubtedly, many of these proteins and peptides are small molecular weight frag- ments of larger signaling molecules. It is now widely recognized that low molecular weight proteins and nuclear fragments can play a role in upregulating important cel- lular growth factors that can lead to skin healing (20–23). EXPERIMENTAL PEPTIDE ISOLATION The peptide used in the following studies was isolated and identifi ed from a low molecu- lar weight fraction of a ferment of S. cerevisiae (24). The peptide amino acid sequence was determined by Erdman Degradation at the University of California, Davis Molecular Structure Facility and indicated that the peptide was a hexapeptide comprised of Phenyl- alanine (Phe), Valine (Val), Alanine (Ala) and Proline (Pro), comprising the unique sequence Phe-Val-Ala-Pro-Phe-Pro (FVAPFP) (Figure 1). The peptide has been assigned the International Nomenclature Cosmetic Ingredient name, Hexapeptide-11. BLAST2® (Washington University, St Louis, MO) available with the National Center Biotechnology Information (NCBI) Sequence Viewer software (http://www.yeastgenomeorg/cgi-bin/ blast-sgd.pl) was employed to match this peptide sequence against the entire protein dataset for S. cerevisiae. The sequence of amino acids that comprise Hexapeptide-11 appears in a number of yeast proteins in particular, the amino acid sequence for Hexapeptide-11 can be found in stress-related proteins (hsp70), and transmembrane proteins as well as a number of proteins whose functions are presently unknown. For the purposes of these studies, the hexapeptide was synthesized using solid state peptide synthesis techniques to a purity of 95% as determined by high-performance liquid chromatography (not shown). The peptide used in the following studies was the highly purifi ed synthetic peptide and all concentrations shown are on a dry basis. Figure 1. Structure of Hexapeptide-11.
JOURNAL OF COSMETIC SCIENCE 82 FIBROBLASTS STUDIES DNA microarray. Human dermal fi broblasts were obtained from Cascade Biologics (Grand Island, NY), seeded into T-25 fl asks, and grown at 37 ± 2°C and 5 ± 1% CO2. Upon reaching confl uency, the cells were treated with Hexapeptide-11 for 24 h after which total RNA was isolated using an RNAqueous Kit (Ambion Inc., Austin, TX) per the manu- facturer’s instructions. After purifi cation, the total RNA was prepared for array use by fi rst amplifying the RNA using a MessageAmp aRNA Kit (Ambion), and then fl uores- cently labeling the aRNA with Cy3 or Cy5 using an ASAP Labeling Kit (Perkin Elmer, Boston, MA), both per the manufacturer’s instructions. To purify the fl uorescently la- beled aRNA, a microcon YM-30 fi lter column was inserted into a collection tube and fi lled with 400 μl of TE buffer. The Cy3 and Cy5 probes were combined and then added to the microcon fi lter and thoroughly mixed with the TE buffer. The fi lter was centri- fuged at 12,000 rpm for 8 min and the fl ow-through was discarded. The column was then washed twice with 400 μl of TE buffer, discarding the fl ow-through each time. After the fi nal wash, the fi lter column was inverted, placed into a new collection tube, and centri- fuged at 2000 rpm for 2 min to collect the probe. The fl uorescently labeled aRNA was applied to the DNA microarray chips (Agilent Technologies, Santa Clara, CA) and the chip was hybridized overnight and washed per the manufacturer’s recommended protocol. After washing, the microarrays were scanned with an Axon GenePix 4100A Scanner (Molecular Devices, Sunnyvale, CA) with the scanning resolution set to 5 μm and analyzed with GenePix Pro software. During the initial scan, the PMT gains for the scanner were adjusted such that the Cy5/ Cy3 image count ratios were between 0.88 and 1.12. Fluorescence intensities for the microarrays were subjected to global normalization. The total fl uorescent signal for both dyes was normalized with a correction factor that would make the ratio of total intensities for both dyes equal to one. For this study, a Cy3/Cy5 (untreated/treated) fl uorescence intensity ratio greater than 1.3 or less than 0.7 (this re- lates to a change in gene expression of at least ±30%) was used as the cutoff for up- and downregulated genes, respectively (25). In addition, the fl uorescence intensity of the gene marker had to be greater than the background intensity. Fibroblast SIPS. For the fi broblast SIPS experiment, fi broblasts were prepared in the same fashion noted above for the array work. The cells were treated for 24 h with the hexapep- tide at various concentrations between 0.01% and 2.0% except the untreated control cells which were treated only with normal cell culture media. At the end of the 24-h in- cubation, the cells were exposed to a sublethal dose of hydrogen peroxide (H2O2) (150 μM, diluted in cell culture media) for 2 h (26). Following the H2O2 exposure period, the media was removed and replaced with fresh, normal cell culture media and the cells were allowed to grow for an additional 24 h after which changes in ATM expression were as- sessed using an immunoblotting-based technique. Fibroblast intrinsic replicative senescence. Human neonatal fi broblasts were obtained after primary culture (passage 1), seeded into a set of T-75 fl asks in 3 ml/fl ask of fi broblast growth media, and grown at 37 + 2°C and 5 + 1% CO2. The cells were expanded through six passages (one passage was defi ned as growing the cells until the fl ask was confl uent and then splitting the cells 1:2, thus one passage was roughly equal to one population doubling). After the 6th pas- sage, the fi broblasts were split into different treatment groups and treated with the various test materials through passage 18. At passage 18, a portion of the fi broblasts were used to assay
Purchased for the exclusive use of nofirst nolast (unknown) From: SCC Media Library & Resource Center (library.scconline.org)