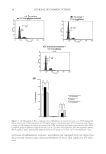
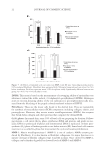
J. Cosmet. Sci., 65, 25–36 (January/February 2014) 25 Molecular marker approaches for tracking redox damage and protection in keratins JOLON M. DYER, CHARISA D. CORNELLISON, ANITA J. GROSVENOR, STEFAN CLERENS, and SANTANU DEB-CHOUDHURY, Food & Bio-Based Products, AgResearch, Christchurch 8140, New Zealand (J.D., C.C., A.G., S.C., S.D.), Riddet Institute at Massey University, Palmerston North, New Zealand (J.D.), and Biomolecular Interaction Centre, University of Canterbury, Christchurch 8140, New Zealand (J.D.). Accepted for publication September 1, 2013. Synopsis There is increasing awareness of the importance of reductive and oxidative (redox) protein damage in protein- based materials including, hair, wool, nails, and skin. Light-induced damage to protein-based materials is of particular concern because of its impact on age-related degradation and product life spans. Consequently, cosmetic applications frequently target hair and skin restoration, where the integrity of the constituent fi la- mentous proteins is essential to a healthy appearance. The keratins constitute an important subset of the structural proteins within skin, hair, and wool. We will introduce a means to assess damage to this important group of proteins at the molecular level, utilizing proteomic techniques to track the formation or degradation of sensitive peptides within intermediate fi lament proteins. The degradation of three molecular markers of redox damage, the peptides SFGYR, LASDDFR, and DVEEWYIR, along with the formation of their oxi- dized products, is demonstrated after exposure to ultraviolet A, ultraviolet B, and blue light. The method is shown to be suitable for evaluating the protective effect of treatments, as lower levels of oxidative markers were observed after the application of a protective fi ber treatment. Molecular-level redox tracking will allow more targeted design and evaluation of protection and repair treatments for protein systems. INTRODUCTION Keratins, previously also referred to as intermediate fi lament proteins, represent an im- portant class of structural proteins of critical relevance to the physico-mechanical proper- ties of mammalian fi bers. Keratins are directly responsible for the structural integrity and associated physical and mechanical properties of hair, wool, skin, and nails (1,2). Both oxidative and reductive damages to these proteins result in deleterious effects, such as loss of strength and an increase in brittleness (3). Address all correspondence to Jolon M. Dyer at jolon.dyer@agresearch.co.nz .
JOURNAL OF COSMETIC SCIENCE 26 Fibrous protein protection and repair technologies are becoming increasingly important in the formulation of functional hair and skin treatments. The evaluation of protein redox damage has traditionally been undertaken at a holistic level, such as through protein ex- traction assays or spectrophotometric carbonyl quantitation (4,5). However, for the tar- geted design, optimization and validation of next-generation protein protection and repair technologies, it is critical that robust and sensitive approaches for the evaluation of protein damage at the molecular level be developed and integrated with analysis at higher structural levels. Here, we present the development and utilization of a peptide-based molecular marker approach for profi ling and tracking oxidative damage in keratins, building on expertise in keratin degradation patterns (6,7). The approach is demonstrated using wool keratins, which have high sequence and structural homology with human hair and therefore repre- sent a good model for evaluating the molecular-level effect of cosmetic treatments on keratins. Redox proteomic approaches were applied to characterize light-induced protein damage at the protein primary level in an enriched keratin extract and to track specifi c modifi cations across a range of irradiation conditions. Molecular markers characterized were then utilized to profi le keratin damage to wool fi bers both with and without the application of a protective silicon matrix fi ber surface treatment, demonstrating the effi - cacy and sensitivity of our approach for evaluating protein protection. MATERIALS AND METHODS Nonylphenol ethoxylate (trade name: Teric GN9) was obtained from Orica Ltd (Mel- bourne, Australia). Ethanol and urea were obtained from Merck Chemicals (Darmstadt, Germany). Thiourea was obtained from Sigma-Aldrich (St. Louis, MO) ChromAR® liquid chromatography (LC)-grade water and LC-grade acetonitrile, sodium tetraborate, and hydrochloric acid from J.T. Baker (Mumbai, India) and Univar formic acid from Ajax Finechem (Waltham, MA). Tris and sodium tetrathionate were obtained from BDH (Poole, UK). Sequencing grade trypsin was obtained from Prom ega (Madison, WI). Acrylamide was obtained from Bio-Rad Laboratories (Hercules, CA). α-Cyano-4- hydroxycinnamic acid was obtained from Bruker Daltonics (Bremen, Germany). Nano- ES spray capillaries were obtained from Proxeon (Odense, Denmark). Wool preparation. Mid-side wool samples from Romney fl eece wool were detipped, then scoured and air-dried. This wool was cut into small sections, then freeze-crushed to a fi ne powder using liquid nitrogen in a mortar and pestle. The crushed wool sample was dried under vacuum over phosphorus pentoxide for 2 days. Whole wool samples were obtained in the form of woven, unbleached Merino wool fabric. Keratin extraction. Dried wool powder was extracted overnight in tubes by vigorous shak- ing in a reciprocal shaker at room temperature in 0.1 M Tris, 0.2 M sodium tetrathionate, 0.1 M sodium tetraborate, and 8 M urea, pH 9.5 (HCl). Tubes were centrifuged to pellet the wool residue. The keratins were purifi ed by precipitation and dialysis, and were then freeze-dried. Keratin purity was assessed by 2D gel electrophoresis, with no other wool protein classes observed. Whole wool surface photoprotection treatment. Solgels were prepared as follows: Ethanol (6 ml) was added to 4.48 ml tetraethoxysilane and stirred for 30 min. Phenyltriethoxysilane (4.48 ml) was added, then 0.8 ml water was added dropwise, followed by 0.3 ml 10 M
Purchased for the exclusive use of nofirst nolast (unknown) From: SCC Media Library & Resource Center (library.scconline.org)