MOLECULAR MARKER APPROACHES FOR TRACKING REDOX DAMAGE 33 Monoisotopic m/z Charge Sequence Modifi cation Protein ID Type II K85 1179.57 2+ KYEQEVALR Nitration (Y) Protein ID Type II K83 1179.58 2+ RYEEEVALR Hydroxylation (Y) Protein ID Type II K81, K86 1195.57 2+ RYEEEVALR Dihydroxylation (Y) Protein ID Type II K81, K86 1240.62 2+ TKYETEVSLR Hydroxylation (Y) Protein ID Type I K33a Phenylalanine 822.38 2+ LAADDFR Hydroxylation (F) Protein ID Type I K31, K32, K33b, K35, K38 838.37 2+ LAADDFR Dihydroxylation (F) Protein ID Type I K31, K32, K33b, K35, K38 1037.50 2+ AKLAADDFR Dihydroxylation (F) Protein ID Type I K31, K33b, K35 Others 719.33 1+ NAQCVK Deamidation (N), carbamidomethyl (C) Protein ID Type II K81, K83, K85, K86 972.49 2+ LLEGQEQR Deamidation (Q) Protein ID Type II K81 974.49 2+ LTAEVENAK Deamidation (N) Protein ID Type II K81, K83, K86 984.43 2+ EHVEADGGR Oxidation (H) Protein ID Type II K85 (Bos taurus) 999.56 1+ LVVQIDNAK Deamidation (N) Protein ID Type I K31, K33a, K33b, K35, Type I K33a 1031.51 2+ QEEKEQIK Deamidation (Q) Protein ID Type II K81, K83, K86 1141.65 2+ LIHEINFLK Oxidation (H) Protein ID Type II K82 (Bos taurus) 1243.64 2+ QLVESDINGLR Deamidation (N) Protein ID Type I K31, K33b 1366.71 2+ LASELNHVQEVL Oxidation (H) Protein ID Type II K87 1399.74 2+ QLVESDINGLRR Deamidation (N) Protein ID Type I K31, K33b 1624.82 2+ LNVEVDAAPPVDLNK Hydroxylation (P) Protein ID Type I K36 (Bos taurus) Backbone cleavage 839.33 1+ DVEEWY None Protein ID Type I K31K33a, K33b, K34 854.34 1+ AEAESWY None Protein ID Type II K81, K83, K85, K86, K87 Table VI Continued
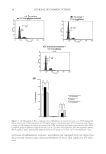
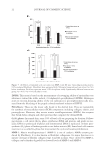
JOURNAL OF COSMETIC SCIENCE 34 hair, and nails, and keratin marker peptides therefore represent an excellent target for proteomic evaluation of damage and protection at the molecular level. For comparison purposes, the yellowness of whole wool irradiated with our protocols was evaluated, and the Y-Z color scores are summarized in Table V. Wool fabric was fi rst irradiated with UVA for 72 h, and a complete redox proteomic profi le compiled through LC-MS/MS analysis. In the case of wool irradiation, modifi cations associ- ated with coloration changes are of particular interest, and so all oxidative modifi cations to the aromatic amino acid residues were identifi ed. The full modifi cation list is shown in Table VI. This proteomic profi ling enabled full profi ling of protein oxidative damage throughout the wool keratins, with characterization of specifi c higher abundance modifi cations for tracking. An ESI-MS/MS-based proteomic evaluation was then performed with and without the application of a protective solgel surface treatment to the wool prior to 48 h UVA and UVB irradiation. Photomodifi cations corresponding to [+O], [+2O], and [+3O] for ty- rosine, and [+O] and [+2O] for tryptophan in keratin marker peptides were tracked proteomically. Figures 3 and 4 show the observed results for the two keratin marker pep- tides, SFGYR and DVEEWYIR. Figure 3. Comparative tracking of the levels of UVA- and UVB-induced modifi cations to tyrosine (Y→[Y+O], [Y+2O], and [Y+3O]) in the marker peptide SFGYR (m/z 629.3). The relative percentage indi- cates the peak area of SFG[Y+O]R, SFG[Y+2O]R, or SFG[Y+3O]R relative to the SFGYR peak area in MS. Triplicate technical repeats were performed, with standard deviation indicated. Figure 4. Comparative tracking of the levels of UVA- and UVB-induced modifi cations to tryptophan (W→[W+O] and [W+2O]) in the marker peptide DVEEWYIR (m/z 1109.6). The relative percentage indi- cates the peak area of DVEEW[W+O]YIR or DVEEW[W+2O]YIR relative to DVEEWYIR peak area in MS. Triplicate technical repeats were performed, with standard deviation indicated.
Purchased for the exclusive use of nofirst nolast (unknown) From: SCC Media Library & Resource Center (library.scconline.org)