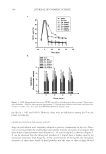
REGULATION OF ECM REMODELING BY XANTHOHUMOL 127 EXPRESSION OF FIBRILLAR COLLAGENS, ELASTIN, AND FIBRILLINS Dermal fi broblasts were cultured in complete Dulbecco’s Modifi ed Eagle’s Medium (DMEM) supplemented with 10% heat-inactivated fetal bovine serum, 1% penicillin/ streptomycin, and 1% L-glutamine (Sigma). For the experiments, cells were trypsinized, seeded in 33-mm dishes, transfected with COL1A1 promoter (or elastin promoter), fi refl y luciferase (pGL4 vector), and TK-hRenilla luciferase plasmids with escort, and dosed with or without (control) xanthohumol (0.01%, 0.1%, 1%) or 2.5 mM ascor- bic acid for 24 hours in basal media containing 1% serum replacement (Sigma) (19–21). The cells were examined for viability by MTS assay (Promega). The media were exam- ined for collagen (types I, III, and V) and elastin fi ber components (tropoelastin, fi brillin-1, and fi brillin-2) with respective antibodies by indirect ELISA (19–21). For ELISA, ali- quots of media (from experiments) or respective standards were added to 96-well plates and incubated overnight at 4°C. The wells were blocked with bovine serum albumin and then incubated with the respective antibodies for one hour at room temperature. The plates were washed thoroughly with wash buffer, incubated with secondary antibody linked to peroxidase for one hour at room temperature, washed with wash buffer thoroughly, and subsequently incubated with peroxidase substrate until the development of color, which was measured spectrophotometrically at 405 nm. The data were normalized for cell via- bility. The cells were lysed and measured for luminescence from fi refl y and renilla luciferase activities with specifi c substrates (Promega). The luminescence from fi refl y luciferase was normalized with that from renilla luciferase. DATA ANALYSIS The effects of xanthohumol or positive controls are represented as a percent of control (no additives) represented at 100%. The data were statistically analyzed for signifi cant differ- ences by ANOVA and Student t-tests at the 95% confi dence interval. Signifi cant effects of xanthohumol, relative to control, are represented by * in the fi gures. RESULTS AND DISCUSSION INHIBITION OF ELASTASE AND MMP-1, 2, AND 9 ACTIVITIES BY XANTHOHUMOL The ECM damaging enzymes are produced by epidermal keratinocytes, dermal fi bro- blasts, and neutrophils, which are the predominant source for photoaged skin (9–14). While collagen is degraded by MMP-1 and MMP-2, the elastic fi bers are degraded by elastase, MMP-9, and MMP-2 (7–9). Xanthohumol inhibited the activities of elastase and MMP-9 at all concentrations (0.001–1%), and MMP-1 and MMP-2 at the higher concentrations (0.1% and 1%), suggesting a greater benefi t to the elastin fi bers (Figure 1a–d). There was complete inhibition of each of the enzyme activities by 1% xanthohu- mol. The concentrations of xanthohumol that inhibited elastase, MMP-9, MMP-1, and MMP-2 activities by 50% were extrapolated as 0.001%, 0.05%, 0.25%, and 0.5%, re- spectively.
JOURNAL OF COSMETIC SCIENCE 128 Relative to the control (0 at 100%), xanthohumol at 0.001%, 0.01%, 0.1%, and 1%, respectively, signifi cantly inhibited elastase activity to 50%, 48%, 23%, and 0.2% of control and inhibited MMP-9 activity to 76%, 68%, 26%, and 3% of control (p0.05) (Figure 1a,d). Xanthohumol at 0.1%, and 1% signifi cantly inhibited MMP-1 activity to 84% and 4% of control, and MMP-2 activity to 70% and –17% of control, respectively (p0.05) (Figure 1b,c). The effects of xanthohumol at the higher concentrations were similar to those of the positive controls ascorbic acid, PMSF or EDTA, and protease Figure 1. Inhibition of elastase (a), MMP-1 (b), MMP-2 (c), and MMP-9 (d) activities by 0.001-1% xan- thohumol (xan within arrows, a-d), PMSF (a), EDTA (b-d), ascorbic acid (AA, a-d), or protease inhibitor (PI, a, b, d). The effect of additives is represented as % of control (0, no additives), represented at 100% *=p0.05, relative to control. Error bars represent standard deviation (n=4).
Purchased for the exclusive use of nofirst nolast (unknown) From: SCC Media Library & Resource Center (library.scconline.org)