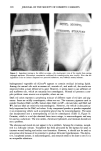
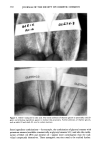
350 JOURNAL OF THE SOCIETY OF COSMETIC CHEMISTS STERILIZATION TIMES A modified preservative efficacy test was used to determine sterilization times (STs) for the test organisms in aqueous samples. The inocula were prepared as above and added to solutions of MP, Na2EDTA, MP + Na2EDTA, and deionized water (control). The contents of the tubes were mixed using a Vortex Genie Mixer, and samples were taken at 0, 2, 4, 24, and 48 hr by inserting a sterile swab into the liquid in each tube. A separate Petri dish containing TSALT was streaked with each swab. The Petri dishes were incubated for 48 hr at 30 ø or 37øC, depending on the test organism, as described above. The Petri dishes were examined for growth of the test organisms, and the ST was determined to be the first time at which test organisms were not recovered from the test solution. The ST and the concentration of organisms in the inoculum were used to calculate the slope of the survivor curve, correcting for the volume change that occurs when the inoculum is added to the test system. In these studies, the concentration of organisms in test tube samples was 1/100th the concentration in the inoculum because 0.1 ml was added to 10-ml solution in each test tube. D-values were determined by taking the negative reciprocal of the slope of each survivor curve (8). Where no endpoint was reached in the ST experiments, because the test organisms were still alive at 48 hr, the minimum possible ST (MPST) was used. The MPST was defined as a time longer than the last time at which test organisms were recovered (i.e., 48 hr). The MPST and the concentration of the organisms in the inoculum were used to construct a virtual survival curve. The maximum possible slope (MPSlope) of the virtual survivor curve and the corresponding minimum possible D-value (MPD-value) were calculated. DETERMINATION OF SYNERGY Synergism was observed when the slope of the survivor curve obtained with the com- bined components was a larger negative number than the sum of the slopes (or MPSlopes) for each of the components determined separately. WATER HARDNESS Duplicate samples of tap water tap water containing 0.01% Na2EDTA, adjusted to pH 7.0 by the addition of one drop of TEA and deionized water were tested for water hardness by the method of Betz Laboratories (9). A 0.1% 1342 dispersion was prepared by slowly adding 0.1 g 1342 to 99.9 g tap water with vigorous agitation. This dispersion was stirred for 2 hr at room temperature to allow hydration of the 1342. The beakers containing tap water and the 1342 dispersion were covered with aluminum foil and were allowed to stand, undisturbed, at room temperature for 3 days. The dispersion settled to about 1/3 of the liquid level in the beaker after this period. The water layer was decanted to give 1342-treated tap water. Duplicate samples of tap water, 1342-treated tap water, and freshly drawn deionized water were assayed for hardness (9). STATISTICS Mean D-values and standard deviation (s) were calculated. Statistically significant dif- ferences between mean D-values of duplicate experiments were determined by a two-
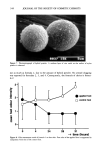
SYNERGY OF PRESERVATIVES 3 51 tailed t test (10). The Q test was used for rejection of a questionable result when five or more D-value determinations were made using the same test organism and test sample (11). RESULTS The survivor curves for P. aeruginosa in 0.2% MP adjusted to pH 7.0 with TEA or NaOH, with and without 0.2% 1342, are shown in Figure 1. The system with 1342, MP, and TEA inactivated P. aeruginosa so quickly that the APC was 600/ml immedi- ately after inoculation. No organisms were recovered at 2 hr or thereafter in this system. The estimated D-value for P. aeruginosa was 0.006 hr, based on the APC of the inoculum and the APC immediately after inoculation. This is indicated by the dashed line in the figure. The system containing MP and 1342 neutralized with NaOH had an initial APC of 4.3 x 105/ml and a D-value of 0.9 hr. Linear regression analysis gave an estimated ST of slightly greater than 5 hr. Solutions of MP and 0.16% NaOH or 0.6% TEA (the amounts of these bases required to adjust the pH to 7.0) did not kill P. aeruginosa during the 24-hr test period. This initial experiment was performed once using duplicate Petri dishes. The results in Table II show the preservative efficacy test results of the nonionic lotion challenged with S. aureus, B. cereus, E. coli, and different species of Pseudomonas. Most species of Pseudomonas were inactivated rapidly, with D-values of • 1.1 hr. The preser- vative system was much less effective against S. aureus, B. cereus, and E. coli than against most species of Pseudomonas tested. Pseudomonas sp., P. cepacia 13945, and P. cepacia 25416 were more resistant than the other pseudomonads to the anti-Pseudomonas action :::) 8 uJ g 1 0 0 5 10 15 20 25 HOUFI8 Figure 1. Survivor curves for P. aeruginosa 9027 in 0.2% MP adjusted to pH 7.0 with TEA or NaOH, with and without 0.2% 1342. Explanation of symbols: ß = 0.2% MP with NaOH, ß = 0.2% MP with TEA, ß = 0.2% MP + 0.2% 1342 with NaOH, and ß = 0.2% MP + 0.2% 1342 with TEA.
Purchased for the exclusive use of nofirst nolast (unknown) From: SCC Media Library & Resource Center (library.scconline.org)