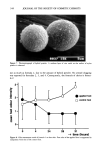
SYNERGY OF PRESERVATIVES 361 Analyses of tap water and 1342-treated tap water indicated that exposure of the water to the 1342 caused a significant decrease in hardness, expressed as ppm CaCO 3. This suggests that 1342 is capable of chelating Ca 2+ ions. The 0.1% 1342 and 0.01% Na2EDTA reduced water hardness by similar amounts. This suggests that these com- pounds have similar chelating abilities for the Ca 2 + ions. The ST study revealed little antibacterial activity by either MP or Na2EDTA alone (Table V). Rapid killing occurred in the presence of MP + Na2EDTA because no viable organisms were recovered at 4 hr in most test systems. The test organisms found to be more persistent in these tests were, in general, more persistent in lotions con- taining 1342 and MP (Table II). In some cases, it is believed that differences in results between these two tables may be attributed to differences in APCs of the inocula. The STs and MPSTs in Table V were used to calculate the slopes and D-values (or MPSlopes and MPD-values) in Table VI. Survivor curve slopes may be determined when the STs and initial inocula of the test organisms are known. For example, ?. aeruginosa 9027 had an ST of 4 hr in MP + Na2EDTA, and the APC in the sample was 1.1 X 105/ml. Here, the D-value and slope were 0.8 hr and - 1.26 hr- •, respectively. Where STs are not known (i.e., ST 48 hr), the MPD-values and corresponding MPSlopes may be estimated from a virtual survivor curve constructed using the APC of the inoculum and the MPST, as explained above. Here, the MPD-value for P. aeruginosa 9027 in MP was calculated to be 9.5 hr and the MPSlope was - 0.105 hr- 1. This slope is the negative reciprocal of the MPD-value and represents the fastest possible rate of death of this organism in this test system. If P. aeruginosa were being killed at a faster rate, then no organisms would have been recovered at the last sampling (i.e., at 48 hr). The MPSlope for P. aeruginosa 9027 in Na 2 EDTA was estimated similarly to be - 0. 105 hr-1 Synergy was observed here, because the slope for the system con- taining MP + Na2EDTA (- 1.26 hr-1) was a larger negative number than the sum of the MPSlopes for MP and for Na2EDTA (- 0.210 hr-1). This procedure was used for each test organism shown in Table VI. The MP q- Na2EDTA system had synergistic anti-Pseudomonas activity for all pseudomonads, except for P. cepacia 13945. The MPD- values for different strains of P. aeruginosa were slightly different due to the slightly different concentrations of organisms in the inocula (Table VI). The estimated STs for S. aureus and E. coli were 48 hr in all test systems consequently, it was not possible to establish synergy for these organisms in this experiment. Numerous workers have reported the enhancement of preservative action by EDTA (3,7,31-34). The potentiation by EDTA is believed to be due to permeabilization synergy, in which one antimicrobial agent (EDTA) assists the passage of the other antimicrobial through the cell wall or membrane (7). We propose that the anti-Pseudo- toohas synergy observed with 934, 941, or 1342 and MP is due, at least in part, to chelation of divalent metal ions and that it is similar to permeabilization synergy re- ported for the potentiation of preservative action by EDTA (7). Results in support of this are the demonstration that 1342 has chelation activity, the elimination of the synergism observed with the fluorescent pseudomonads by the addition of 0.1% CaC12 (Table IV), and the similarities in the survival patterns of the various pseudomonads in nonionic lotions with polyacrylic acid or acrylic acid copolymer/MP systems (Table II) and in Na2EDTMMP solutions (Table V). Similar patterns of inactivation were observed in both aqueous and nonionic emulsion
362 JOURNAL OF THE SOCIETY OF COSMETIC CHEMISTS systems. This suggests that the observed synergy is not due to the nonionic emulsions studied here. The effect of 934, 941, or 1342 and MP on the inactivation of Pseudomonas was not tested in anionic or cationic emulsion systems or in various surfactant systems such as anionic shampoos and liquid soaps. The reason for this is that 1342 emulsion systems are sensitive to electrolytes, which cause loss of emulsion stability (35). Adair et al. (36) reported that P. aeruginosa 9027 underwent lysis following metabolism of di- or tricarboxylic acids and sodium lauryl sulfate, and that lysis was not due to chelation. It is evident that the mechanism reported by these workers is not the same as the mechanism observed in the current work. The antibacterial synergy of MP and acrylic acid homopolymer/copolymers against most Pseudomonas test cultures was not observed with E. coli 8739. This gram-negative or- ganism was not inactivated rapidly in nonionic emulsions containing MP and 1342 or in solutions containing MP + Na2EDTA (Tables II, V, and VI). These findings sug- gest a mechanism of action that is relatively specific for pseudomonads and not other gram-negative bacteria however, testing with other strains of E. coli and other gram- negative organisms is necessary to confirm this. The survivor curve slope method of determining synergy of preservative systems has application to both current experimental findings and to data presented in the litera- ture. Application of this method to the ST data of Richards and Hardie (37) revealed synergism for fentichlor/phenylethanol combinations against E. coli and Proteus vulgaris at 0.0015% or 0.0050% fentichlor q- 0.4% phenylethanol, and with 0.0050% fen- tichlor + 0.4% phenylethanol against P. aeruginosa. The survivor curve slope method revealed increased antibacterial activity against S. aureus,' however, this effect was not synergistic. Our use of the survivor curve slope method corroborated the findings of Richards and Hardie. The survivor curve slope method of determining synergy was applied to the D-values for S. aureus, in various combinations of preservatives in saline, reported by Akers et al. (38). Slopes of the survival curves were determined by taking the negative reciprocal of the D-values reported by these workers, and these slopes were used to determine whether mixtures of two preservatives exhibited synergy. Although these workers did not attempt to determine synergy, application of the survival curve slope method to their data for linear analysis of preservatives in saline solutions revealed that systems containing 0.2% phenol q- 0.3% m-cresol, 0.2% phenol q- 0.2% m-cresol, and 0.2% MP + 1.0% benzyl alcohol exhibited synergy. Akers et al. ranked the efficacy of these preservative systems in the top half of the systems tested against S. aureus. The survival curve slope method was used to study synergy in a system reported by Boehm to be synergistic (4). He reported that 0.25% P and 0.09% N were synergistic against P. pyocyanea. Since we did not have the same strain as in Boehm's experiments, it was decided to "bracket" the concentrations of P and N used in Boehm's studies. The results in Table VII show the growth response observed with P. aeruginosa 9027 in 0 to 1% P and/or 0 to 0.1% N. The STs and MPSTs in Table VII were used to calculate the slopes, MPSlopes, D-values, and MPD-values in Table VIII. Our findings show syn- ergy for concentrations that bracket the synergistic combination reported by Boehm. The use of kinetic parameters--the slopes of survivor curves obtained by use of the linear regression method--to demonstrate anti-Pseudomonas synergy of MP and acrylic acid homopolymer/copolymers in vitro has not been reported previously. The survivor
Purchased for the exclusive use of nofirst nolast (unknown) From: SCC Media Library & Resource Center (library.scconline.org)