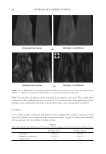
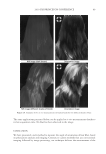
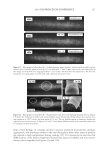
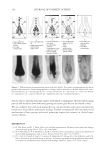
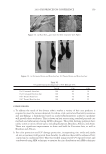
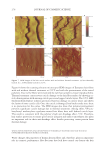
2010 TRI/PRINCETON CONFERENCE 233 characterizing bases for the defects. AKR mice, for example, display hair with an “interior defect” consisting of haphazard patterns of medulla cell arrangement due to mutations in steroyl O-acetyltransferase 1 (24). In strains without this defect, where medulla cells are typically in an orderly alignment with regular spacing, electron microscopy of mature hair shafts reveals projections from the cortex into the middle of each medulla cell. The AKR strain lacks these indentations, evidently permitting irregular or disorderly spacing of the medulla cells. Proteomic analysis revealed only a low level of trichohyalin, a major compo- nent of the projections, in the cortical cells of AKR mice compared to two other strains not showing this phenotype (4). In the course of this work, it was noted that all three mouse strains could be distinguished by their proteomic profi les, raising the question whether, by analogy, human hair from individuals of different ethnic origin could be distinguished by protein profi ling. This approach to analysis of hair is easily adaptable to other keratinized structures such as nail plate. The nail plate proteome resembles that of the hair shaft in its high content of keratin proteins and the more complex profi le of the insoluble fraction compared to the solubilized proteins (25). Nearly all the 30 proteins in the solubilized fraction were kera- tins and keratin-associated proteins, while the insoluble fraction was comprised of, in addition to these, cytoplasmic proteins, membrane and junctional proteins and histones (Figure 3). Many of the proteins overlapped considerably with those detected in hair shaft, notably some abundant keratins (K31, K33B, K34, K39, K85, K86), junctional proteins (DSP, JUP, DSG4, PKP1) and numerous intracellular proteins (SELENBP1, SFN, ACTB, HSPA5). Some proteins were detected only in nail plate, including certain keratins (K5, K6A, K14, K17), junctional proteins (DSG1, EPPK1) and intracellular proteins (KPRP, SERPINB12, CKB). Others seen only in hair shaft included several keratins (K40, K82) and intracelluar proteins (CTTNB1, ATP5B). The fi nding of keratins (and KPRP) found only in nail, which are also found in epidermis, supports the established view that the nail unit expresses features of both hair and epidermis (26). Figure 3. Cellular locations of identifi ed proteins in nail plate sorted into the categories keratin and keratin- associated proteins (Ker), other cytoplasmic proteins (Cyt), membrane and junctional proteins (Mem), and histones (His). Relative amounts were estimated based on unique peptides by the exponentially modifi ed protein abundance index (21,25).
JOURNAL OF COSMETIC SCIENCE 234 INTERPRETATION AND PROSPECTS The nail unit and hair follicle produce structures that are remarkably tough and cohesive by virtue of their protein components, an abundance of intramolecular and intermolecular disul- fi de bonds and translgutaminase-mediated isopeptide bonding connecting the intermediate fi lament matrix to the cell surface. The fi nal stage of the intricate keratinocyte differentiation program involves transglutaminase activation by increasing concentration of intracellular cal- cium ions, thereby connecting membrane and junctional proteins to the cytoskeleton. The enzymes encoded by the TGM1 and TGM3 genes appear effective in that most of the proteins found in the solubilized fraction are also found in the insoluble (cross-linked) fraction of both hair shaft and nail plate. That so many proteins from all the compartments of the cell were identifi ed in the insoluble fraction is evidence that the corneocyte incorporates available pro- tein rather than a few specifi c proteins. A future goal for hair is to analyze the proteome of cuticle, cortex and medulla cells separately. This has now been accomplished for wool cuticle, where 100 proteins were identifi ed representing a variety of cellular processes (27). Hair shaft, nail plate and epidermal callus, the latter through sampling with tape strips, provide essentially noninvasive sources of discrete protein subsets of the total organismal proteome. The shotgun approach to analysis is even anticipated to permit distinguishing the proteomes of epidermis at various anatomic sites, including glabrous surfaces or those infl uenced by adjoining abnormal conditions (e.g., acne). For analysis of disease states, it has the advantage of surveying many gene products in parallel, permitting discovery of single components that may be defi cient. While homozygous protein loss may be readily detect- able, the approach does have obvious limitations for identifying the basis for any given disease or adverse condition, since only the most prominent proteins are surveyed. Even if a given protein is identifi ed in the sample examined, a point mutation could easily be over- looked if the protein coverage did not include the peptide in which the mutation occurs, or the affected peptide, one of many unique peptides, is not specifi cally monitored. Neverthe- less, downstream effects of a given defect might still be visible in altered levels of other proteins that may be important for the phenotype. Detection of a heterozygous defective allele in a carrier of a recessive condition is also problematic without better quantitation. Complementing the above shotgun (or discovery) approach, targeting specifi c peptides has potential utility. For example, quantitating only a small number of proteotypic peptides from a given protein, those unique to that protein and obtained reproducibly in high yield with suitable fragmentation patterns, can provide improved relative quantitation (28). This approach (multiple reaction monitoring) would be attractive if it would permit de- tection of heterozygous gene loss. For those diseases where a limited number of mutations were anticipated, and they occurred in suitable locations in a detectable protein, expected mutant peptides could be monitored. Additionally, focusing on specifi c peptides has the advantage of permitting much greater sensitivity. Ultimately, a desirable outcome would be to use a panel of specifi c peptides to distinguish alternative diagnoses for given diseases or conditions. While this would not replace the gold standard of genetic testing, it could serve as a useful screen in a comprehensive diagnostic paradigm. ACKNOWLEDGMENTS This work was supported by USPHS grant P42 ES04699 from the National Institute of Environmental Health Sciences. I thank Drs. Young Jin Lee, Rich Eigenheer and Brett
Purchased for the exclusive use of nofirst nolast (unknown) From: SCC Media Library & Resource Center (library.scconline.org)