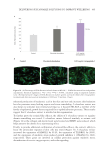
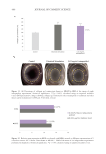
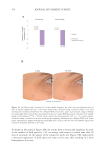
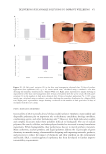
503 Evolution and Challenges of Sustainability
their gene/RNA sequences. The archaea are not known to cause skin infections, disease, or
product contamination consequently, they are not discussed in detail in this review.
In 2018, Byrd, Belkaid, and Segre studied bacteria on different anatomical sites using 16S
rRNA sequencing methodology.74 They reported the 10 most abundant bacteria found
in dry sites (e.g., hypothenar palm and volar forearm), moist sites (e.g., nares, antecubital
fossa, inguinal crease, interdigital web, and popliteal fossa), sebaceous sites (e.g., alar crease,
cheek, glabella, external auditory canal, manubrium, retroauricular crease, occiput, and
back), and the foot (e.g., toe web space, toenail, and plantar heel). The species identified are
listed in Table VI.
Fungi are also members of the skin microbiome and generally comprise members of the
Ascomycota and Basidiomycota phyla.74 The predominant genus is Malassezia spp. (formerly
Pityrosporum spp.), which grow in sebum rich regions of the skin, such as the scalp, face and
upper back) and have been associated with dandruff and seborrheic dermatitis. Byrd and
coworkers reported that the highest level of fungal diversity occurs on the feet, which
frequently are colonized by Aspergillus, Cryptococccus, Rhodotorula, and Epicoccum genera.
Byrd and coworkers also studied fungi on different anatomical sites and identified them
using the internal transcribed spacer 1 (ITS1) region of the eukaryotic ribosomal gene for
sequencing. They reported the 10 most abundant fungi found in the same four skin sites
as were used for the bacteria (above). The 10 most abundant fungal species identified are
listed in Table VII. The differences in the findings between Grice and coworkers and Byrd
and coworkers may be due to factors including different panelists, demographics, season of
the year, and methodology.
Most viruses on skin are bacteriophages (i.e., phages), which are viruses that infect and
replicate within bacteria and archaea. Natarelli, Gahoonia, and Sivamani observed that
viral diversity present in chronic wounds is increased compared to healthy skin.75 A study
conducted using an atopic mouse model showed favorable results for the use of phage
Table VI
Top 10 Abundant Bacterial on Various Skin Sites
Dry Moist Sebaceous Foot
Cutibacterium acnes Corynebacterium
tuberculostericum
Cutibacterium acnes Corynebacterium
tuberculostericum
Corynebacterium
tuberculostericum
Staphylococcus hominis Staphylococcal epidermidis Staphylococcus
hominis
Streptococcus mitis Cutibacterium acnes Corynebacterium
tuberculostericum
Staphylococcus
warneri
Streptococcus oralis Staphylococcus epidermidis Staphylococcus capitis Staphylococcus
epidermidis
Streptococcus pseudopneumoniae Staphylococcus capitis Corynebacterium stimulans Staphylococcus capitis
Streptococcus sanguinis Corynebacterium fastidiosum Streptococcus mitis Staphylococcus
haemolyticus
Micrococcus luteus Corynebacterium afermentans Staphylococcus hominis Micrococcus luteus
Staphylococcus epidermidis Micrococcus luteus Corynebacterium aurimucosum Corynebacterium
afermentans
Staphylococcus capitis Enhydrobacter aerosaccus Corynebacterium kroppenstedtii Corynebacterium
simulans
Veillonella parvula Corynebacterium simulans Corynebacterium amycolatum Corynebacterium
resistens
*Table adapted from Byrd, Belkaid, and Segre.74
their gene/RNA sequences. The archaea are not known to cause skin infections, disease, or
product contamination consequently, they are not discussed in detail in this review.
In 2018, Byrd, Belkaid, and Segre studied bacteria on different anatomical sites using 16S
rRNA sequencing methodology.74 They reported the 10 most abundant bacteria found
in dry sites (e.g., hypothenar palm and volar forearm), moist sites (e.g., nares, antecubital
fossa, inguinal crease, interdigital web, and popliteal fossa), sebaceous sites (e.g., alar crease,
cheek, glabella, external auditory canal, manubrium, retroauricular crease, occiput, and
back), and the foot (e.g., toe web space, toenail, and plantar heel). The species identified are
listed in Table VI.
Fungi are also members of the skin microbiome and generally comprise members of the
Ascomycota and Basidiomycota phyla.74 The predominant genus is Malassezia spp. (formerly
Pityrosporum spp.), which grow in sebum rich regions of the skin, such as the scalp, face and
upper back) and have been associated with dandruff and seborrheic dermatitis. Byrd and
coworkers reported that the highest level of fungal diversity occurs on the feet, which
frequently are colonized by Aspergillus, Cryptococccus, Rhodotorula, and Epicoccum genera.
Byrd and coworkers also studied fungi on different anatomical sites and identified them
using the internal transcribed spacer 1 (ITS1) region of the eukaryotic ribosomal gene for
sequencing. They reported the 10 most abundant fungi found in the same four skin sites
as were used for the bacteria (above). The 10 most abundant fungal species identified are
listed in Table VII. The differences in the findings between Grice and coworkers and Byrd
and coworkers may be due to factors including different panelists, demographics, season of
the year, and methodology.
Most viruses on skin are bacteriophages (i.e., phages), which are viruses that infect and
replicate within bacteria and archaea. Natarelli, Gahoonia, and Sivamani observed that
viral diversity present in chronic wounds is increased compared to healthy skin.75 A study
conducted using an atopic mouse model showed favorable results for the use of phage
Table VI
Top 10 Abundant Bacterial on Various Skin Sites
Dry Moist Sebaceous Foot
Cutibacterium acnes Corynebacterium
tuberculostericum
Cutibacterium acnes Corynebacterium
tuberculostericum
Corynebacterium
tuberculostericum
Staphylococcus hominis Staphylococcal epidermidis Staphylococcus
hominis
Streptococcus mitis Cutibacterium acnes Corynebacterium
tuberculostericum
Staphylococcus
warneri
Streptococcus oralis Staphylococcus epidermidis Staphylococcus capitis Staphylococcus
epidermidis
Streptococcus pseudopneumoniae Staphylococcus capitis Corynebacterium stimulans Staphylococcus capitis
Streptococcus sanguinis Corynebacterium fastidiosum Streptococcus mitis Staphylococcus
haemolyticus
Micrococcus luteus Corynebacterium afermentans Staphylococcus hominis Micrococcus luteus
Staphylococcus epidermidis Micrococcus luteus Corynebacterium aurimucosum Corynebacterium
afermentans
Staphylococcus capitis Enhydrobacter aerosaccus Corynebacterium kroppenstedtii Corynebacterium
simulans
Veillonella parvula Corynebacterium simulans Corynebacterium amycolatum Corynebacterium
resistens
*Table adapted from Byrd, Belkaid, and Segre.74