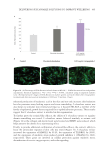
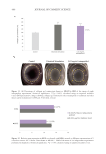
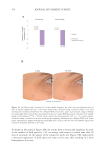
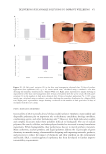
502 JOURNAL OF COSMETIC SCIENCE
by regulating temperature, preventing water loss from the body, reducing irritation/
injury due to harsh chemicals and harmful radiation, enabling touch and pain sensations,
supporting vitamin D synthesis, and preventing infection. The skin surface and appendages
are colonized by a diverse community of microorganisms, the “skin microbiome,” and
these microorganisms are involved with maintaining the skin ecosystem with their lipid
metabolism, colonization resistance to transient and pathogenic organisms, regulation of
the skin immune system (SIS), and creation of acid mantle condition of skin.66-69
The skin microbiome is composed of bacteria, yeasts, fungi, viruses, micro-eukaryotes
(mites), and archaea. Although the average number of microorganisms isolated from skin
using traditional culture methods (e.g., plating on agar media) ranges from 103–104 cfu/cm2,
humid places on the body, such as armpits, groin, and nostrils, may have microbial counts
exceeding 106 cfu/cm2. The microbial counts on the scalp, forehead and around the ears are
about 106 cfu/cm2, while microbial counts on the upper back, chest and arms range from
104–105 cfu/cm2.67,69,70 In 2016, Patra. Byrne and Wolf reported that 23–100% of healthy
people have dust mites, which generally are found on the face, around sebaceous glands,
and in hair follicles.71 Consequently, they are part of the normal microflora. Demodex mites
(Demodex folliculorum and Demodex brevis) are present on skin and may be associated with
rosacea and blepharitis.71
In the 2009 landmark publication by Grice et al., it was reported that skin sustains
microorganisms that influence human health and disease.72 They observed that traditional
culture-based characterizations of the skin microflora are biased toward species that readily
grow under standard laboratory conditions, such as the staphylococci, and that molecular
approaches have revealed a greater diversity of skin microflora in distinct topographical
regions of the skin. This provided the basis for examining multiple skin sites with the use
of genomic techniques.
Grice and coworkers analyzed 16S ribosomal RNA (rRNA) gene sequences from 20 distinct
skin sites of 10 healthy human test subjects and found that comparable sites on different
people harbor similar bacterial communities. Although the number of people being tested
was rather small, it was determined that 90% of the bacteria on skin could be classified
into four types (phyla): Actinobacteria (52% Gram positive bacteria with high (55%)
guanosine plus cytosine (G+C) DNA that include Corynebacterium, Actinomyces, Arthrobacter,
Micrococcus, Mycobacterium, Nocardia, Propionibacterium (Cutibacterium) and Rhodococcus
genera), Firmicutes (24% Gram positive bacteria with low G+C DNA content that include
Staphylococcus, Lactobacillus, Streptococcus, Clostridium and Bacillus genera), Proteobacteria
(16% Gram-negative bacteria that include Escherichia, Pseudomonas, Salmonella, Vibrio and
Helicobacter genera) and Bacteroidetes (6% Gram-negative rod-shaped bacteria that include
Bacteroides and Porphyromonas genera). Grice and coworkers found that coagulase negative
staphylococci (CNS) such as Staphylococcus epidermidis, along with Cutibacterium acnes
(formerly Propionibacterium acnes), Corynebacterium spp., Micrococcus spp., Streptococcus spp.,
and Acinetobacter spp. were the dominant genera present on skin.72 Many of bacteria in these
genera were found in studies of skin microflora reported by Samaras and Hoptroff in which
they estimated that Cutibacterium, Staphylococcus, and Corynebacterium genera constituted up
to 80% of the entire skin microbiome.73
Grice and coworkers noted that about 4% of the skin microbiome is represented by the
archaea, based on 16S rRNA sequencing.72 Archaea is a domain of single-celled organisms
that lack cell nuclei and are therefore prokaryotic like bacteria. However, most archaea have
not been isolated by traditional microbiological methods and have been detected only by
by regulating temperature, preventing water loss from the body, reducing irritation/
injury due to harsh chemicals and harmful radiation, enabling touch and pain sensations,
supporting vitamin D synthesis, and preventing infection. The skin surface and appendages
are colonized by a diverse community of microorganisms, the “skin microbiome,” and
these microorganisms are involved with maintaining the skin ecosystem with their lipid
metabolism, colonization resistance to transient and pathogenic organisms, regulation of
the skin immune system (SIS), and creation of acid mantle condition of skin.66-69
The skin microbiome is composed of bacteria, yeasts, fungi, viruses, micro-eukaryotes
(mites), and archaea. Although the average number of microorganisms isolated from skin
using traditional culture methods (e.g., plating on agar media) ranges from 103–104 cfu/cm2,
humid places on the body, such as armpits, groin, and nostrils, may have microbial counts
exceeding 106 cfu/cm2. The microbial counts on the scalp, forehead and around the ears are
about 106 cfu/cm2, while microbial counts on the upper back, chest and arms range from
104–105 cfu/cm2.67,69,70 In 2016, Patra. Byrne and Wolf reported that 23–100% of healthy
people have dust mites, which generally are found on the face, around sebaceous glands,
and in hair follicles.71 Consequently, they are part of the normal microflora. Demodex mites
(Demodex folliculorum and Demodex brevis) are present on skin and may be associated with
rosacea and blepharitis.71
In the 2009 landmark publication by Grice et al., it was reported that skin sustains
microorganisms that influence human health and disease.72 They observed that traditional
culture-based characterizations of the skin microflora are biased toward species that readily
grow under standard laboratory conditions, such as the staphylococci, and that molecular
approaches have revealed a greater diversity of skin microflora in distinct topographical
regions of the skin. This provided the basis for examining multiple skin sites with the use
of genomic techniques.
Grice and coworkers analyzed 16S ribosomal RNA (rRNA) gene sequences from 20 distinct
skin sites of 10 healthy human test subjects and found that comparable sites on different
people harbor similar bacterial communities. Although the number of people being tested
was rather small, it was determined that 90% of the bacteria on skin could be classified
into four types (phyla): Actinobacteria (52% Gram positive bacteria with high (55%)
guanosine plus cytosine (G+C) DNA that include Corynebacterium, Actinomyces, Arthrobacter,
Micrococcus, Mycobacterium, Nocardia, Propionibacterium (Cutibacterium) and Rhodococcus
genera), Firmicutes (24% Gram positive bacteria with low G+C DNA content that include
Staphylococcus, Lactobacillus, Streptococcus, Clostridium and Bacillus genera), Proteobacteria
(16% Gram-negative bacteria that include Escherichia, Pseudomonas, Salmonella, Vibrio and
Helicobacter genera) and Bacteroidetes (6% Gram-negative rod-shaped bacteria that include
Bacteroides and Porphyromonas genera). Grice and coworkers found that coagulase negative
staphylococci (CNS) such as Staphylococcus epidermidis, along with Cutibacterium acnes
(formerly Propionibacterium acnes), Corynebacterium spp., Micrococcus spp., Streptococcus spp.,
and Acinetobacter spp. were the dominant genera present on skin.72 Many of bacteria in these
genera were found in studies of skin microflora reported by Samaras and Hoptroff in which
they estimated that Cutibacterium, Staphylococcus, and Corynebacterium genera constituted up
to 80% of the entire skin microbiome.73
Grice and coworkers noted that about 4% of the skin microbiome is represented by the
archaea, based on 16S rRNA sequencing.72 Archaea is a domain of single-celled organisms
that lack cell nuclei and are therefore prokaryotic like bacteria. However, most archaea have
not been isolated by traditional microbiological methods and have been detected only by