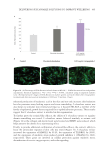
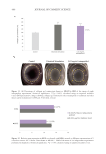
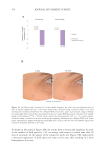
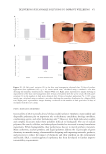
518 JOURNAL OF COSMETIC SCIENCE
(63) Ciccognani DT. Cosmetic preservation using enzymes. In: Orth DS, Kabara JJ, Denyer SP, Tan SK, eds.
Cosmetic and Drug Microbiology. Informa Healthcare USA, Inc 2006:185-203.
(64) Stiles ME. Bacteriocins produced by Leuconostoc species. J Dairy Sci. 1994 77(9):2718-2724. doi:10.3168/
jds.S0022-0302(94)77214-3
(65) Zhong X, Wang G, Li F, et al. Immunomodulatory effect and biological significance of β-glucans.
Pharmaceutics. 2023 15(6):1615. doi:10.3390/pharmaceutics15061615
(66) Skowron K, Bauza-Kaszewska J, Kraszewska Z, et al. Human skin microbiome: impact of intrinsic and
extrinsic factors on skin microbiota. Microorganisms. 2021 9(3):543. doi:10.3390/microorganisms9030543
(67) Gallo RL. Human Skin is the largest epithelial surface for interaction with microbes. J Invest Dermatol.
2017 137(6):1213-1214. doi:10.1016/j.jid.2016.11.045
(68) Dimitriu PA, Iker B, Malik K, Leung H, Mohn WW, Hillebrand GG. New insights into the intrinsic and
extrinsic factors that shape the human skin microbiome. mBio. 2019 10(4):1-14. doi:10.1128/mBio.00839-19
(69) Cundell AM. Microbial ecology of the human skin. Microb Ecol. 2018 76(1):113-120. doi:10.1007/
s00248-016-0789-6
(70) Andersen BM. Prevention and Control of Infections in Hospitals: Practice and Theory. 1st ed Springer Nature
2018:337-437.
(71) Patra V, Byrne SN, Wolf P. The skin microbiome: is it affected by UV-induced immune suppression?
Front Microbiol. 2016 7:1235. doi:10.3389/fmicb.2016.01235
(72) Grice EA, Kong HH, Conlan S, et al. Topographical and temporal diversity of the human skin
microbiome. Science. 2009 324(5931):1190-1192. doi:10.1126/science.1171700
(73) Samaras S, Hoptroff M. The microbiome of healthy skin. In: Dayan N, ed. Skin Microbiome Handbook:
from Basic Research to Product Development. 1st ed. Scrivener Publishing LLC 2020:3-32.
(74) Byrd AL, Belkaid Y, Segre JA. The human skin microbiome. Nat Rev Microbiol. 2018 16(3):143-155.
doi:10.1038/nrmicro.2017.157
(75) Natarelli N, Gahoonia N, Sivamani RK. Bacteriophages and the microbiome in dermatology: the
role of the phageome and a potential therapeutic strategy. Int J Mol Sci. 2023 24(3):2095. doi:10.3390/
ijms24032695
(76) Shimamori Y, Mitsunaka S, Yamashita H, et al. Staphylococcal phage in combination with Staphylococcus
epidermidis as a potential treatment for Staphylococcus aureus-associated atopic dermatitis and
suppressor of phage-resistant mutants. Viruses. 2020 13(1):7. doi:10.3390/v13010007
(77) Barnard E, Shi B, Kang D, Craft N, Li H. The balance of metagenomic elements shapes the skin
microbiome in acne and health. Sci Rep. 2016 6:39491. doi:10.1038/srep39491
(78) Orth DS. Normal microflora of human skin. Insights into cosmetic microbiology. Allured Bus Media.
2010:211-239.
(79) Swaney MH, Kalan LR. Living in your skin: microbes, molecules, and mechanisms. Infect Immun.
2021 89(4):1-18. doi:10.1128/IAI.00695-20
(80) Fournière M, Latire T, Souak D, Feuilloley MGJ, Bedoux G. Staphylococcus epidermidis and
Cutibacterium acnes: Two major sentinels of skin microbiota and the influence of cosmetics.
Microorganisms. 2020 8(11):1752. doi:10.3390/microorganisms8111752
(81) Orth DS. The probiotic nature of normal microflora. Cosmet Toiletr. 2008 123(8):41,42:44-46.
(82) Flowers L, Grice EA. The skin microbiota: balancing risk and reward. Cell Host Microbe. 2020 28(2):190-
200. doi:10.1016/j.chom.2020.06.017
(83) Borkowski AW, Gallo RL. The coordinated response of the physical and antimicrobial peptide barriers
of the skin. J Invest Dermatol. 2011 131(2):285-287. doi:10.1038/jid.2010.360
(84) Kiatsurayanon C, Ogawa H, Niyonsaba F. The role of host defense peptide human β-defensins in the
maintenance of skin barriers. Curr Pharm Des. 2018 24(10):1092-1099. doi:10.2174/13816128246661803
27164445
(85) Becknell B, Spencer JD. A Review of Ribonuclease 7’s Structure, Regulation, and Contributions to Host
Defense. Int J Mol Sci. 2016 17(3):423. doi:10.3390/ijms17030423
(63) Ciccognani DT. Cosmetic preservation using enzymes. In: Orth DS, Kabara JJ, Denyer SP, Tan SK, eds.
Cosmetic and Drug Microbiology. Informa Healthcare USA, Inc 2006:185-203.
(64) Stiles ME. Bacteriocins produced by Leuconostoc species. J Dairy Sci. 1994 77(9):2718-2724. doi:10.3168/
jds.S0022-0302(94)77214-3
(65) Zhong X, Wang G, Li F, et al. Immunomodulatory effect and biological significance of β-glucans.
Pharmaceutics. 2023 15(6):1615. doi:10.3390/pharmaceutics15061615
(66) Skowron K, Bauza-Kaszewska J, Kraszewska Z, et al. Human skin microbiome: impact of intrinsic and
extrinsic factors on skin microbiota. Microorganisms. 2021 9(3):543. doi:10.3390/microorganisms9030543
(67) Gallo RL. Human Skin is the largest epithelial surface for interaction with microbes. J Invest Dermatol.
2017 137(6):1213-1214. doi:10.1016/j.jid.2016.11.045
(68) Dimitriu PA, Iker B, Malik K, Leung H, Mohn WW, Hillebrand GG. New insights into the intrinsic and
extrinsic factors that shape the human skin microbiome. mBio. 2019 10(4):1-14. doi:10.1128/mBio.00839-19
(69) Cundell AM. Microbial ecology of the human skin. Microb Ecol. 2018 76(1):113-120. doi:10.1007/
s00248-016-0789-6
(70) Andersen BM. Prevention and Control of Infections in Hospitals: Practice and Theory. 1st ed Springer Nature
2018:337-437.
(71) Patra V, Byrne SN, Wolf P. The skin microbiome: is it affected by UV-induced immune suppression?
Front Microbiol. 2016 7:1235. doi:10.3389/fmicb.2016.01235
(72) Grice EA, Kong HH, Conlan S, et al. Topographical and temporal diversity of the human skin
microbiome. Science. 2009 324(5931):1190-1192. doi:10.1126/science.1171700
(73) Samaras S, Hoptroff M. The microbiome of healthy skin. In: Dayan N, ed. Skin Microbiome Handbook:
from Basic Research to Product Development. 1st ed. Scrivener Publishing LLC 2020:3-32.
(74) Byrd AL, Belkaid Y, Segre JA. The human skin microbiome. Nat Rev Microbiol. 2018 16(3):143-155.
doi:10.1038/nrmicro.2017.157
(75) Natarelli N, Gahoonia N, Sivamani RK. Bacteriophages and the microbiome in dermatology: the
role of the phageome and a potential therapeutic strategy. Int J Mol Sci. 2023 24(3):2095. doi:10.3390/
ijms24032695
(76) Shimamori Y, Mitsunaka S, Yamashita H, et al. Staphylococcal phage in combination with Staphylococcus
epidermidis as a potential treatment for Staphylococcus aureus-associated atopic dermatitis and
suppressor of phage-resistant mutants. Viruses. 2020 13(1):7. doi:10.3390/v13010007
(77) Barnard E, Shi B, Kang D, Craft N, Li H. The balance of metagenomic elements shapes the skin
microbiome in acne and health. Sci Rep. 2016 6:39491. doi:10.1038/srep39491
(78) Orth DS. Normal microflora of human skin. Insights into cosmetic microbiology. Allured Bus Media.
2010:211-239.
(79) Swaney MH, Kalan LR. Living in your skin: microbes, molecules, and mechanisms. Infect Immun.
2021 89(4):1-18. doi:10.1128/IAI.00695-20
(80) Fournière M, Latire T, Souak D, Feuilloley MGJ, Bedoux G. Staphylococcus epidermidis and
Cutibacterium acnes: Two major sentinels of skin microbiota and the influence of cosmetics.
Microorganisms. 2020 8(11):1752. doi:10.3390/microorganisms8111752
(81) Orth DS. The probiotic nature of normal microflora. Cosmet Toiletr. 2008 123(8):41,42:44-46.
(82) Flowers L, Grice EA. The skin microbiota: balancing risk and reward. Cell Host Microbe. 2020 28(2):190-
200. doi:10.1016/j.chom.2020.06.017
(83) Borkowski AW, Gallo RL. The coordinated response of the physical and antimicrobial peptide barriers
of the skin. J Invest Dermatol. 2011 131(2):285-287. doi:10.1038/jid.2010.360
(84) Kiatsurayanon C, Ogawa H, Niyonsaba F. The role of host defense peptide human β-defensins in the
maintenance of skin barriers. Curr Pharm Des. 2018 24(10):1092-1099. doi:10.2174/13816128246661803
27164445
(85) Becknell B, Spencer JD. A Review of Ribonuclease 7’s Structure, Regulation, and Contributions to Host
Defense. Int J Mol Sci. 2016 17(3):423. doi:10.3390/ijms17030423